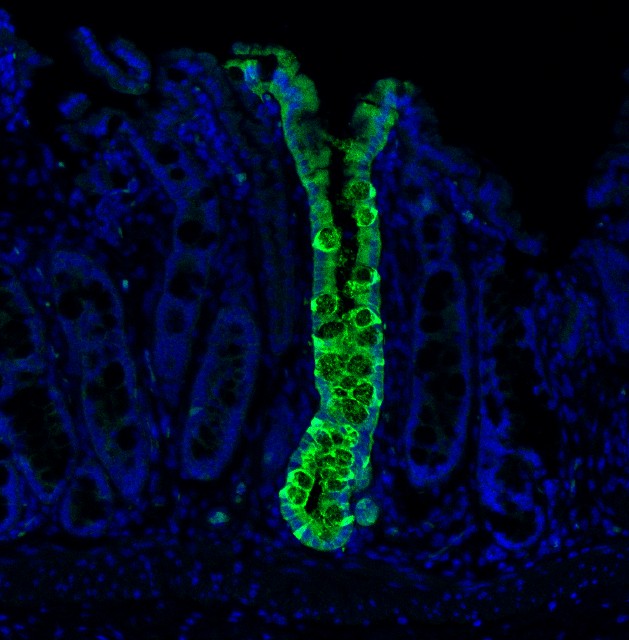
The accurate evaluation of cell identity has long represented a challenge due to the lack of any systematic means to assess the fidelity of engineered cells. We developed CellNet, a network biology-based computational platform that accurately evaluates cell fate through gene regulatory network reconstruction and generates hypotheses for improving cell differentiation protocols. Using this platform, we surveyed a range of engineered cells and found that cells derived via directed differentiation more faithfully recapitulated target cell identity than cells generated by direct conversion. These directly converted cells commonly failed to silence expression programs of the original cell type, and illicit gene expression programs frequently surfaced. Employing induced hepatocytes generated from fibroblasts as a prototypical conversion, our computational and functional analyses showed that iHeps behave as embryonic progenitors with the potential to functionally engraft both the liver and colon. We found that these engineered cells resembled mature colonic epithelium after transplantation into the colon niche, leading us to rename this cell type, ‘induced endoderm progenitors’ (iEPs).
Recently, we have revealed further mechanisms of reprogramming to iEPs. Successful reprogramming is a rare event; thus, it had remained a challenge to isolate and analyze the few iEPs emerging from fibroblasts. High-throughput single-cell RNA-sequencing has enabled this heterogeneity to be deconstructed, although lineage relationships between cells were lost; interpretation of the resulting data presented a challenge. To overcome this, we have developed a straightforward, high-throughput cell tracking method, CellTagging. Sequential lentiviral delivery of heritable random unique molecular indexes, CellTags, permits the construction of multi-level lineage trees. Application of CellTagging to direct lineage reprogramming to iEPs has allowed us to define successful reprogramming trajectories and enhance the yield of iEPs.
Overall, we apply our technologies to study gene regulatory networks, to dissect and engineer cell fate of clinically relevant tissues. This focus integrates three major themes: First, we aim to understand how transcription factor overexpression drives changes in the transcriptional program to remodel cell identity, and how we can exploit this to derive desired cell types. Second, we transplant engineered cells into the in vivo niche, tracking their maturation to understand the steps required to fully differentiate cells in vitro. Finally, we employ single-cell transcriptomics to understand how cell fate is reprogrammed and matured, formulating a blueprint of cell identity to help engineer fate in vitro. Ultimately, we wish to translate new insights in cell fate specification into better human models of disease and eventually into the development of novel therapeutic strategies.
Visit ‘research’ for more information.
Reprogramming has generated a lot of clinical interest. Many diseases, such as Alzheimer’s, diabetes, and heart failure, arise as a result of absent or malfunctioning cells. Readily available cells, such as those from the skin or blood, can be reprogrammed back to embryonic stages. In the petri dish, we can then mimic natural development to coax them to become medically useful cell types, such as cells of the heart or liver. Ultimately we’d like to replace a patient’s diseased tissues with these cells, but for now, they are proving very useful for studying human disease and development in the dish.
While this avenue holds a lot of promise, the cell types generated are often immature and lack full adult function. Also, it is a complicated process to reprogram the cells and then mature them. There is an alternative approach, though. Rather than using the Yamanaka factors to deliver cells to an embryonic state, we can use other gene combinations to change cellular identity. This method, designed to avoid immature stages, is called ‘direct conversion’ and aims to directly transform one adult cell type into another mature adult cell type. This method has been thought to be faster and more efficient than full reprogramming. Our research found that direct conversion only partially converts cells, and returns them to later embryonic stages. Only when transplanted into a living animal do the cells mature into their fully functioning adult state.
Our lab aims to engineer cell identity, focusing on medically relevant tissues to repair/replace diseased and damaged organs. One way in which we manipulate fate is to artificially switch on genes in cells, observing how this controls cell identity. To further understand this, we also look at how genes are turned on/off during developmental stages. In this respect, the embryo provides a blueprint of identity. Finally, we contribute to the Human Cell Atlas consortium to make a high-resolution map of every cell type in the human body. Ultimately we aim to create cells for transplant into patients, but we can also use these engineered cells as a valuable tool to study disease and development in the dish.
The lab is situated in the Longwood Medical Area, Boston, USA. We are members of the Divisions of GI and Genetics at Brigham and Women’s Hospital and Harvard Medical School. The Principal Investigator of the lab, Dr. Samantha Morris, is originally from the United Kingdom. She completed her Ph.D. at the University of Cambridge, where she remained to investigate early mammalian development in the lab of Magdalena Zernicka-Goetz. Samantha then relocated to the USA in 2011 to work on cell fate engineering in the lab of George Daley at Boston Children’s Hospital and Harvard Medical School. The Morris lab opened in July 2015 with a focus on combining these areas of expertise in stem cell and developmental biology, and single-cell technology development, to generate clinically valuable cell types for disease modeling and regenerative therapy.
Visit ‘Lab Members’ for more information.
Contact
Boston, MA 02115